Transforming Data¶
Overview¶
SolveBio makes it easy to transform data using its dynamic, Python-based expression language. You can use expressions to transform data when importing files, when copying data between (or within) datasets (using "migrations"), or even when querying datasets. In all scenarios, expressions can be provided through the target_fields parameter. To learn more about expressions, please visit the Expressions section.
This article describes how to transform data using dataset migrations, but you can use the same techniques with dataset imports. With dataset migrations, you can copy data between datasets as well as modify datasets in-place. This makes it possible to add, edit, and remove fields. All dataset migrations have a source dataset and a target dataset (which can be the same when editing a single dataset).
Copy a Dataset¶
Copying a dataset is the simplest dataset migration. In this case, a new dataset is created (the target) and all the records from a source dataset are copied to it. The source dataset remains unchanged. Copying datasets can be useful if you do not want to alter the source dataset, or if you do not have write access to the source dataset.
If you do not want to copy the entire source dataset, you can provide filter parameters to copy a subset. This example copies all BRCA1 variants from ClinVar into a new dataset:
1 2 3 4 5 6 7 8 9 10 | from solvebio import Dataset # Retrieve the source dataset source = Dataset.get_by_full_path('solvebio:public:/ClinVar/3.7.4-2017-01-30/Variants-GRCh37') # Create your new target dataset target = Dataset.get_or_create_by_full_path('~/python_examples/clinvar') # Copy all variants in BRCA1 query = source.query().filter(gene_symbol='BRCA1') query.migrate(target=target) |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | library(solvebio) # Retrieve the source dataset source_dataset <- Dataset.get_by_full_path('solvebio:public:/ClinVar/3.7.4-2017-01-30/Variants-GRCh37') # Create your new target dataset vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/clinvar", sep=":") target_dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # Copy all variants in BRCA1 migration <- DatasetMigration.create( source_id=source_dataset$id, target_id=target_dataset$id, # Omit source_params to copy the whole dataset source_params=list( filters=list( list('gene_symbol', 'BRCA1') ) ) ) |
Add Fields¶
The most common dataset transformation is to add a field to a dataset (also known as annotating the dataset or inserting a column). Fields can be added or modified using the target_fields parameter, which should contain a list of valid dataset fields. Any new fields in target_fields will be automatically detected and added to the dataset's schema. Adding fields requires the use of the upsert or overwrite commit mode, depending on the desired effect. This will ensure that the records are updated in-place (based on their _id value), and not duplicated.
A collection of fields is a Dataset Template
To add multiple fields and transform data in a specific way, use a Dataset Template
In this example, a new field will is added to a dataset "in-place", using upsert commit mode.
This example follows from the previous one
To run this example, make sure you have created your own copy of ClinVar.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | from solvebio import Dataset dataset = Dataset.get_or_create_by_full_path('~/python_examples/clinvar') fields = [ { 'name': 'clinsig_clone', 'expression': 'record.clinical_significance', 'description': 'A copy of the current clinical_significance value', 'data_type': 'string' } ] # The source and target are the same dataset, which edits the dataset in-place dataset.migrate(target=dataset, target_fields=fields, commit_mode='upsert') |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/clinvar", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) fields = list( list( name='clinsig_clone', expression='record.clinical_significance', data_type='string' ) ) # The source and target are the same dataset, which edits the dataset in-place DatasetMigration.create( source_id=dataset$id, target_id=dataset$id, target_fields=fields, source_params=list(limit=100000), commit_mode='upsert' ) |
Recipes¶
A field can also be added using a recipe, which is the UI equivalent of inserting a column.
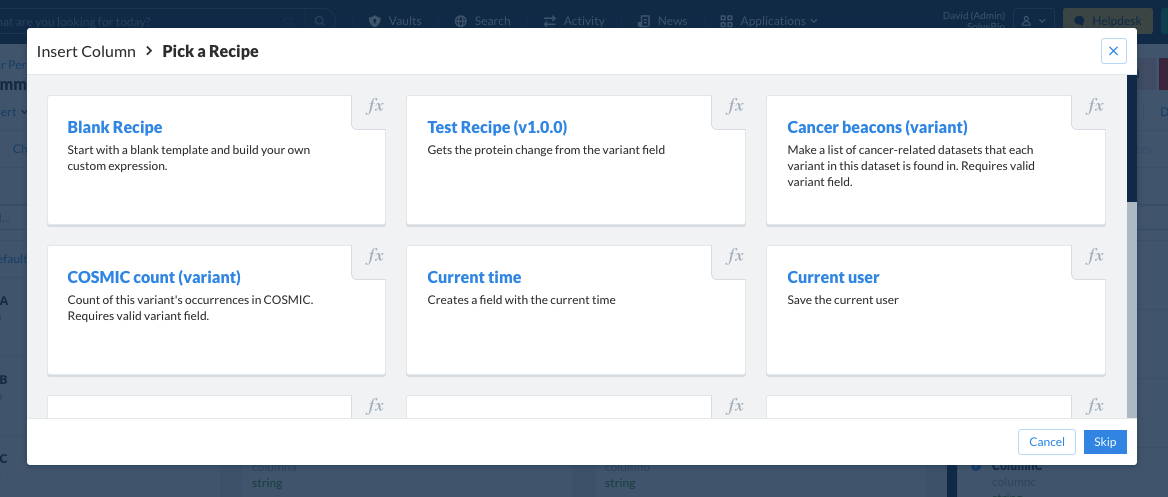
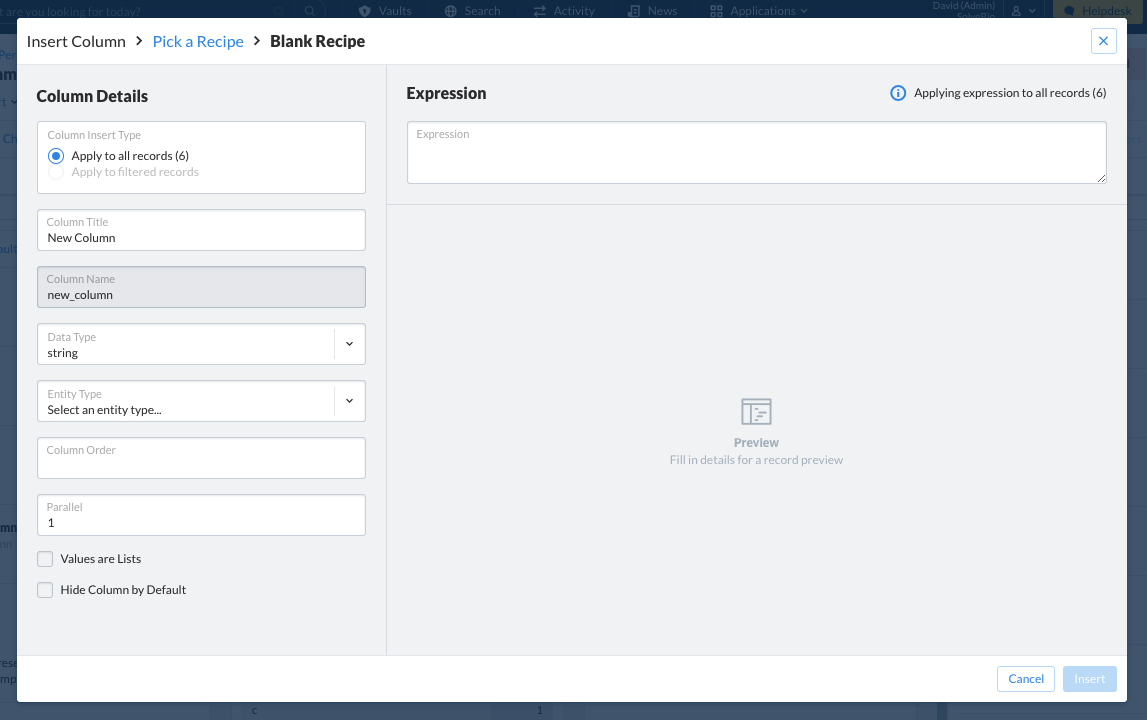
Preset recipes are available for use for commonly created fields. A custom recipe can also be built using a blank recipe and constructing the field and expression manually.
Adding a column that already exists will overwrite data
Note that if the column that is specified already exists in the dataset, data will be overwritten.
Parallel Mode (beta)
NOTE: This feature is currently in testing and only available to some users.
In the UI, an additional integer value called parallel can also be set to split a single migration into multiple parallel tasks, attempting to improve run time. While the value defaults to 1 (run a single task), any integer between 1 and 50 can be chosen.
Edit Fields¶
In this example, an existing field is modified (converted to uppercase). Similar to the example above, a commit mode of overwrite or upsert must be used here to avoid duplicating records.
This example uses an expression that references a pre-existing field in the dataset (learn more about expression context).
This example follows from the previous one
To run this example, make sure you have created your own copy of ClinVar with the new field added.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | from solvebio import Dataset dataset = Dataset.get_or_create_by_full_path('~/python_examples/clinvar') fields = [ { # Convert your copied "clinical_significance" values to uppercase. 'name': 'clinsig_clone', 'data_type': 'string', 'expression': 'value.upper()' } ] # The source and target are the same dataset, which edits the dataset in-place dataset.migrate(target=dataset, target_fields=fields, commit_mode='upsert') |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/clinvar", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) fields <- list( list( # Convert your copied "clinsig_clone" values to uppercase. name= 'clinsig_clone', data_type='string', expression='value.upper()' ) ) # Run a migration where the source and target are the same dataset DatasetMigration.create( source_id=dataset$id, target_id=dataset$id, target_fields=fields, commit_mode='upsert' ) |
Remove Fields¶
Removing a field requires a new empty target dataset
Since you cannot remove a field from a dataset in-place, all the data must be migrated to a new target dataset with the removed field(s) excluded from the source dataset.
In this example, the field (clinsig_clone) in the source dataset is removed by the dataset migration. Since we are removing a field, the target dataset must be a new dataset (or one without the field). In this scenario, any commit mode can be used unless you intend to overwrite records in the target dataset.
This example follows from the previous one
To run this example, make sure you have created your own copy of ClinVar with the new field added.
1 2 3 4 5 6 7 8 9 10 11 | from solvebio import Dataset # Use the dataset from the example above source = Dataset.get_or_create_by_full_path('~/python_examples/clinvar') # To remove a field, you need to create a new, empty dataset first. target = Dataset.get_or_create_by_full_path('~/python_examples/clinvar_lite') # Exclude the copied field from the example above query = source.query(exclude_fields=['clinsig_clone']) query.migrate(target=target, commit_mode='upsert') |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | library(solvebio) vault <- Vault.get_personal_vault() # Use the dataset from the example above source_full_path <- paste(vault$full_path, "/r_examples/clinvar", sep=":") source <- Dataset.get_by_full_path(source_full_path) # To remove a field, you need to create a new, empty dataset first. target_full_path <- paste(vault$full_path, "/r_examples/clinvar_lite", sep=":") target <- Dataset.get_or_create_by_full_path(target_full_path) # Exclude the copied field from the example above DatasetMigration.create( source_id=source$id, target_id=target$id, source_params=list( exclude_fields=list('clinsig_clone') ), commit_mode='upsert' ) |
If you just want to remove the data from a specific field, run an upsert migration and use an expression to set the values to None (Python's equivalent to NULL).
Transient Fields¶
Transient fields are like variables in a programming language. They can be used in a complex transform that requires intermediate values that you do not want to store in the dataset. Transient fields can be referenced by other expressions, but are not added to the dataset's schema or stored. Just add is_transient=True and ensure that the field's ordering evaluates the transient fields in the right order.
In the following example we will use transient fields to structure a few VCF records, leaving SolveBio variant IDs and dbSNP rsIDs in the resulting dataset:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 | from solvebio import Dataset dataset = Dataset.get_or_create_by_full_path('~/python_examples/transient_test') # Sample of a VCF file vcf = """CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA00001 NA00002 NA00003 20 14370 rs6054257 G A 29 PASS NS=3;DP=14;AF=0.5;DB;H2 GT:GQ:DP:HQ 0|0:48:1:51,51 1|0:48:8:51,51 1/1:43:5:.,. 20 17330 . T A 3 q10 NS=3;DP=11;AF=0.017 GT:GQ:DP:HQ 0|0:49:3:58,50 0|1:3:5:65,3 0/0:41:3 20 1110696 rs6040355 A G 67 PASS NS=2;DP=10;AF=0.333,0.667;AA=T;DB GT:GQ:DP:HQ 1|2:21:6:23,27 2|1:2:0:18,2 2/2:35:4 20 1230237 . T T 47 PASS NS=3;DP=13;AA=T GT:GQ:DP:HQ 0|0:54:7:56,60 0|0:48:4:51,51 0/0:61:2 20 1234567 microsat1 GTCT GTACT 50 PASS NS=3;DP=9;AA=G GT:GQ:DP 0/1:35:4 0/2:17:2 1/1:40:3 """.splitlines() # Extract the records (without the header) records = [{'row': row} for row in vcf[1:]] target_fields = [ { "name": "header", "is_transient": True, "ordering": 1, "data_type": "string", "is_list": True, "expression": vcf[0].split() }, { "name": "row", "is_transient": True, "ordering": 1, "data_type": "object", "expression": "dict(zip(record.header, value.split()))" }, { "name": "build", "is_transient": True, "ordering": 1, "data_type": "string", "expression": "'GRCh37'" }, { "name": "variant", "ordering": 2, "data_type": "string", "entity_type": "variant", "expression": """ '-'.join([ record.build, record.row['CHROM'], record.row['POS'], record.row['POS'], record.row['ALT'] ]) """ }, { "name": "rsid", "data_type": "string", "ordering": 2, "expression": "get(record, 'row.ID') if get(record, 'row.ID') != '.' else None" } ] imp = DatasetImport.create( dataset_id=dataset.id, records=records, target_fields=target_fields) dataset.activity(follow=True) for record in dataset.query(exclude_fields=['_id', '_commit']): print(record) # {'rsid': 'rs6054257' , 'variant': 'GRCh37-20-14370-14370-A'} # {'rsid': None , 'variant': 'GRCh37-20-17330-17330-A'} # {'rsid': 'rs6040355' , 'variant': 'GRCh37-20-1110696-1110696-G'} # {'rsid': None , 'variant': 'GRCh37-20-1230237-1230237-T'} # {'rsid': 'microsat1' , 'variant': 'GRCh37-20-1234567-1234567-GTACT'} |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/variants-transient", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # translate_variant() returns an object with 8+ fields. # since we do not want all those fields added to the dataset # we make this field transient and pull the gene and protein change values # into their own fields target_fields <- list( list(name="translated_variant_transient", description="Transient fields that runs variant translation expression", is_transient=TRUE, data_type="object", ordering=1, expression="translate_variant(record.variant)"), list(name="gene", description="HUGO Gene Symbol", data_type="string", ordering=2, expression="get(record, 'translated_variant_transient.gene')"), list(name="protein_change", data_type="string", ordering=2, expression="get(record, 'translated_variant_transient.protein_change')") ) # Import these records into a dataset records <- list( list(variant='GRCH37-17-41244429-41244429-T'), list(variant='GRCH37-3-37089131-37089131-T') ) # Add columns gene/protein_change, via transient column "translated_variant" imp <- DatasetImport.create(dataset_id = dataset$id, records = records, commit_mode = 'upsert', target_fields = target_fields) Dataset.activity(dataset$id) Dataset.query(id = dataset$id, exclude_fields=list('_id', '_commit')) # gene variant protein_change # 1 BRCA1 GRCH37-17-41244429-41244429-T p.S1040N # 2 MLH1 GRCH37-3-37089131-37089131-T p.K618M |
Transforming Queries¶
The output of regular dataset queries can be transformed using the target_fields parameter. This makes it possible to supplement the information returned from a query in real-time, without having to modify the underlying dataset.
The following example retrieves a number of variants from ClinVar and annotates the records with gnomAD population allele frequencies:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 | from solvebio import Dataset target_fields = [{ 'name': 'gnomad_af', 'data_type': 'float', 'expression': """ dataset_field_values( 'solvebio:public:/gnomAD/2.1.1/Exomes-GRCh38', field='af', entities=[('variant', record.variant)]) """ }] dataset = Dataset.get_by_full_path('solvebio:public:/ClinVar/5.1.0-20200720/Variants-GRCH38') query = dataset.query(target_fields=target_fields).filter(gene='BRCA2') for row in query[:10]: print(row['variation_id'], row['variant'], row['gnomad_af']) |
1 2 3 4 5 6 7 8 9 10 11 12 | library(solvebio) dataset <- Dataset.get_by_full_path("solvebio:public:/ClinVar/5.1.0-20200720/Variants-GRCH38") filters <- '[["gene__exact","BRCA2"]]' fields <- list( list( name='gnomad_af', data_type='float', expression='dataset_field_values("solvebio:public:/gnomAD/2.1.1/Exomes-GRCh38", field="af", entities=[("variant", record.variant)])' ) ) query <- Dataset.query(dataset$id, filters=filters, target_fields=fields, limit=10) |
Modifying Datasets¶
Overwrite Records¶
In order to completely overwrite specific records in a dataset just use the overwrite commit mode. You'll need to know the _id of the records you wish to overwrite, but you can get that by querying the dataset.
In this example we will import a few records and then edit one of them:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 | from solvebio import Dataset from solvebio import DatasetImport dataset = Dataset.get_or_create_by_full_path('~/python_examples/edit_records') # Initial Import imp = DatasetImport.create( dataset_id=dataset.id, records=[ {'name': 'Francis Crick', 'birth_year': '1916'}, {'name': 'James Watson', 'birth_year': '1928'}, {'name': 'Rosalind Franklin', 'birth_year': '1920'} ] ) # Retrieve the record to edit record = dataset.query().filter(name='Francis Crick')[0] record['name'] = 'Francis Harry Compton Crick' record['awards'] = ['Order of Merit', 'Fellow of the Royal Society'] # Overwrite mode imp = DatasetImport.create( dataset_id=dataset.id, records=[record], commit_mode='overwrite' ) # Lookup record by ID and see the edited record print(dataset.query().filter(_id=record['_id'])) |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/transform_overwrite", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # Initial records to import records <- list( list(name='Francis Crick', birth_year='1916'), list(name='James Watson', birth_year='1928'), list(name='Rosalind Franklin', birth_year='1920') ) imp <- DatasetImport.create(dataset_id = dataset$id, records = records) # Get record and change some data record <- Dataset.query(id = dataset$id, filters='[["name", "Francis Crick"]]')[1,] record['name'] = 'Francis Harry Compton Crick' record['awards'] = list('Order of Merit', 'Fellow of the Royal Society') # Overwrite mode imp <- DatasetImport.create(dataset_id = dataset$id, records = list(as.list(record)), commit_mode = 'overwrite') # Show the result filters = list( list("_id", record$"_id") ) Dataset.query(id = dataset$id, filters = filters) |
Pro Tip
To clear all values in a field, try setting the expression to None and run the migration.
Upsert (Edit) Records¶
In order to only update (or add) specific fields in a dataset use the upsert commit mode. You'll need to know the _id of the records you wish to upsert, but you can get that by querying the dataset.
Similar to the example above, we will import a few records and then edit one of them:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 | from solvebio import Dataset from solvebio import DatasetImport dataset = Dataset.get_or_create_by_full_path('~/python_examples/upsert_records') # Initial Import imp = DatasetImport.create( dataset_id=dataset.id, records=[ {'name': 'Francis Crick', 'birth_year': '1916'}, {'name': 'James Watson', 'birth_year': '1928'}, {'name': 'Rosalind Franklin', 'birth_year': '1920'} ] ) # Retrieve the record to edit record = dataset.query().filter(name='Francis Crick')[0] # Change existing field record['name'] = 'Francis Harry Compton Crick' # Add a new field record['birthplace'] = 'Northampton, England' # Upsert mode imp = DatasetImport.create( dataset_id=dataset.id, records=[record], commit_mode='upsert' ) # Lookup record by ID and see the edited record print(dataset.query().filter(_id=record['_id'])) |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/transform_upsert", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # Initial records to import records <- list( list(name='Francis Crick', birth_year='1916'), list(name='James Watson', birth_year='1928'), list(name='Rosalind Franklin', birth_year='1920') ) imp <- DatasetImport.create(dataset_id = dataset$id, records = records) # Get record and change some data record <- Dataset.query(id = dataset$id, filters='[["name", "Francis Crick"]]')[1,] record['name'] = 'Francis Harry Compton Crick' record['birthplace'] = 'Northampton, England' # Upsert mode imp <- DatasetImport.create(dataset_id = dataset$id, records = list(as.list(record)), commit_mode = 'upsert') # Show the result filters = list( list("_id", record$"_id") ) Dataset.query(id = dataset$id, filters = filters) |
Delete Records¶
In order to completely delete a record from a dataset you may use the delete commit mode, and pass a list of record IDs (from their _id field).
You can delete records via an import (if you have a file or list of record IDs) or via a migration (if you are deleting the results of a dataset query).
Delete via Import¶
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 | from solvebio import Dataset from solvebio import DatasetImport dataset = Dataset.get_or_create_by_full_path('~/python_examples/data_delete') # Initial Import imp = DatasetImport.create( dataset_id=dataset.id, records=[ {'name': 'six'}, {'name': 'seven'}, {'name': 'eight'}, {'name': 'nine'} ] ) for r in dataset.query(fields=['name']): print(r) # {'name': 'six'} # {'name': 'seven'} # {'name': 'eight'} # {'name': 'nine'} # Get the record ID for 'nine' record = dataset.query(fields=['_id']).filter(name='nine') # Delete the record imp = DatasetImport.create( dataset_id=dataset.id, records=list(record), commit_mode='delete' ) # Why was six afraid of seven? for r in dataset.query(fields=['name']): print(r) # {'name': 'six'} # {'name': 'seven'} # {'name': 'eight'} |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/data_delete", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # Initial records to import records <- list( list(name='six'), list(name='seven'), list(name='eight'), list(name='nine') ) imp <- DatasetImport.create(dataset_id = dataset$id, records = records) Dataset.query(id = dataset$id, fields=list('name')) # name # 1 six # 2 seven # 3 eight # 4 nine # Get the record ID for 'nine' record <- Dataset.query(id = dataset$id, filters='[["name", "nine"]]', fields=list("_id")) # Delete mode imp <- DatasetImport.create(dataset_id = dataset$id, records = list(as.list(record)), commit_mode = 'delete') Dataset.query(id = dataset$id, fields=list('name')) # name # 1 six # 2 seven # 3 eight |
Delete via Migration¶
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 | from solvebio import Dataset from solvebio import DatasetImport dataset = Dataset.get_or_create_by_full_path('~/python_examples/data_delete_migration') # Initial Import imp = DatasetImport.create( dataset_id=dataset.id, records=[ {'name': 'Alice'}, {'name': 'Bob'}, {'name': 'Carol'}, {'name': 'Chuck'}, {'name': 'Craig'}, {'name': 'Dan'}, {'name': 'Eve'} ] ) # Get the records where names begin with C query = dataset.query().filter(name__prefix='C') # Use migration shortcut to Delete migration = query.migrate(dataset, commit_mode="delete") # The above shortcut is equivalent to: # migration = DatasetMigration.create( # source_id=dataset.id, # target_id=dataset.id, # commit_mode="delete", # source_params=dict(filters=[('name__prefix', 'C')]) # ) for r in dataset.query(): print(r) # {'name': 'Alice'}, # {'name': 'Bob'}, # {'name': 'Dan'}, # {'name': 'Eve'} |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 | library(solvebio) vault <- Vault.get_personal_vault() dataset_full_path <- paste(vault$full_path, "/r_examples/data_delete_migration", sep=":") dataset <- Dataset.get_or_create_by_full_path(dataset_full_path) # Initial records to import records <- list( list(name='Alice'), list(name='Bob'), list(name='Carol'), list(name='Chuck'), list(name='Craig'), list(name='Dan'), list(name='Eve') ) imp <- DatasetImport.create(dataset_id = dataset$id, records = records) # Get the records where names begin with C records <- Dataset.query(id = dataset$id, filters='[["name__prefix", "c"]]') # Delete mode migration <- DatasetMigration.create( source_id = dataset$id, target_id = dataset$id, source_params=list(filters=list(list("name__prefix", "C"))), commit_mode = 'delete') Dataset.query(id = dataset$id, fields=list('name')) # name # 1 Alice # 2 Bob # 3 Dan # 4 Eve |