Entities¶
Overview¶
Entities are special labels for dataset fields that contain specific content, such as genes, variants, vault objects, samples, and more. Entities allow for cross-dataset data harmonization, easy filtering, Beacons, and other entity-specific functions.
Supported Entites¶
The following entities are supported:
| Entity | Description | Example |
|---|---|---|
| sample | Sample ID (may also refer to a patient, aliquot, or replicate). This is the basic bio-unit that a set of variants may belong to. | TCGA-02-0001-01 |
| vault_objects | SolveBio vault object ID (can be a file, dataset, or folder) | 510110131292845817 |
| literature | Pubmed ID of a scientific paper | 19684571 |
| genomic_region | Chromosome and start/stop position of a genomic interval | GRCH38-7-117559590-117559593 |
| vault | SolveBio vault ID | 2956 |
| gene | Gene symbol (using standard HUGO nomenclature) | BRCA2 |
| variant | SolveBio variant ID for a unique variant | GRCH38-7-140753336-140753336-T |
| dataset | SolveBio dataset ID | 1126936965182430633 |
| copy_number_variant | SolveBio copy number variant ID | BRCA2 amplification |
| subject | Subject ID | SUBJ001 |
Setting Entities¶
Entities can be set on import or later via the web UI or API.
Automated import¶
SolveBio automatically extracts and labels the right fields as entities for common genomics filetypes such as VCF and GFF3/GTFs. For all other files, SolveBio's entity detection automatically detects if fields contain certain entities such as genes or variants.
Manually on import¶
Entities can be manually set on data import. This can be done via a template on data import for new datasets or data migration for existing datasets. Please see importing data for an example.
Web¶
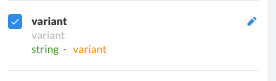
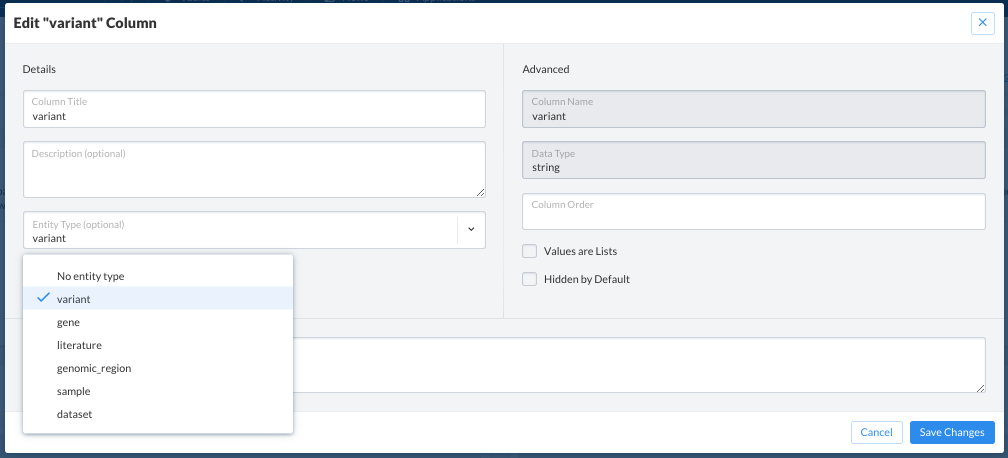
Entities can be added, removed, or switched to any field on SolveBio on any dataset where the user has write access. On the dataset view, any field with a orange label next to the field type is an entity field. Entities can be changed by clicking on the pencil icon.
This opens a modal where the entity can be removed, reset, or added.
Using Entities on SolveBio¶
Explorers¶
Variants, genes, and literature have web explorers for individual entities, which brings together the wealth of public information about the entity, tailored for each type of entity. These explorers also display beacons for each entity, or which public or private datasets where this entity has been found. See examples such as BRCA2, EGFR T790M, and 19684571 for gene, variant, and literature respectively.
Filtering datasets directly¶
Datasets can be queried by entities instead of specifying the exact field name that the entity labels. Expression that query datasets can also use entity filters directly.
Cross-dataset comparisons¶
Many common representations of each entity can be harmonized for easy comparison with the entity_ids expression.
Finally, variant datasets have samples in common, can be used in the Variant Comparison App workflow.