Archiving Datasets¶
Overview¶
Archiving gives you the ability to safely store the datasets that you do not use frequently, without consuming your organization's active storage space quota. When you decide that you want to use the dataset again, you can quickly and easily restore it. Depending on the storage class used, a dataset may be archived automatically.
Permissions¶
A user must have write permissions on the vault in order to archive or restore a dataset.
Querying¶
Archived datasets currently cannot be queried and will raise an error if a query is attempted. You can check if a dataset is archived by checking its availability parameter. The value will be available, unavailable, or archived.
Examples¶
You can easily archive and restore a dataset through the UI or through the API (via Python or R).
SolveBio UI¶
You can archive or restore from the UI in several different places.
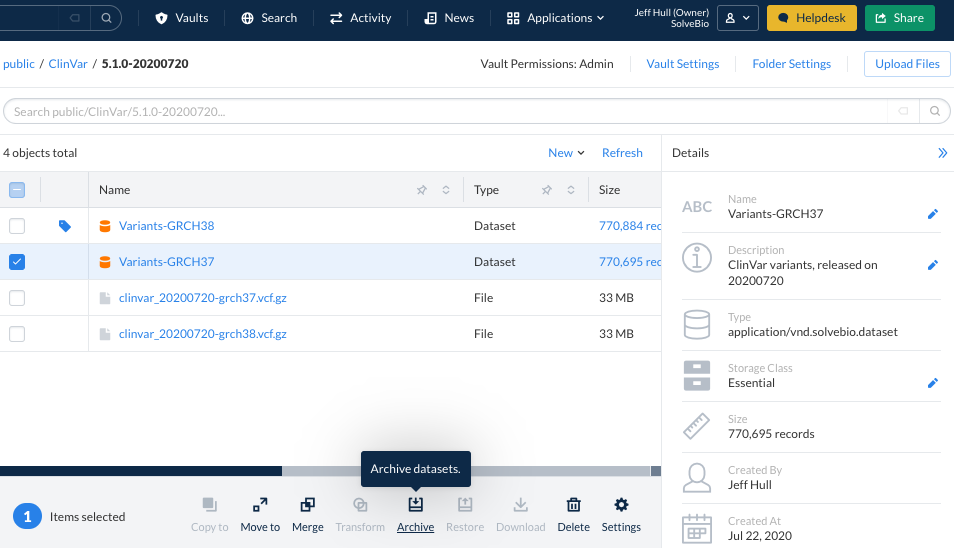
From the vaults view, you can modify a single dataset or multiple datasets at once by selecting from the actions available below. Or you can modify the dataset by clicking on the pencil icon on the right-hand side dataset details pane:
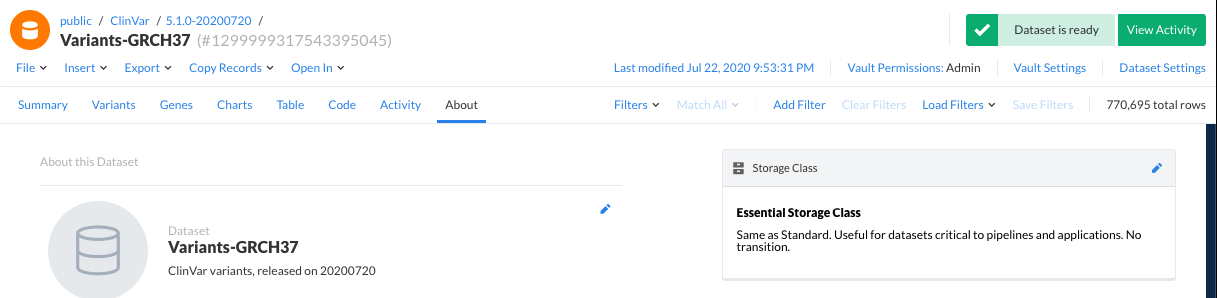
From the dataset view about tab, launch the dataset settings from the top right corner, or by clicking on the pencil icon by the storage class details box. If the dataset has been archived, there will be an explicit "Restore dataset" button.
Archiving¶
A Dataset can be archived using the archive() function within Python, or by changing the storage class to "Archive" within the R client.
1 2 3 4 5 6 7 8 9 10 | import solvebio as sb # Retrieve the dataset by dataset_id dataset = sb.Object.retrieve('dataset_id') dataset.archive() # Archive all datasets in a folder, recursively folder = Object.retrieve('folder_id') for dataset in folder.datasets(recursive=True): dataset.archive() |
1 2 3 4 | require(solvebio) # Set storage class to archive Object.update("DATASET ID", storage_class="Archive") |
Restore¶
Restore of the archived dataset can be done using the restore() function on the archived dataset. By default the Python client will use the "Standard" storage class. However you may restore to any storage class that is available.
1 2 3 4 | import solvebio as sb dataset = sb.Object.retrieve('dataset_id') dataset.restore() |
1 2 3 4 | require(solvebio) # Restore the dataset by setting the storage class to standard Object.update("DATASET ID", storage_class="Standard") |
Switching the Storage Class¶
Storage classes can be modified from the Python/R clients as follows:
1 2 3 4 5 6 7 | import solvebio as sb dataset = sb.Object.retrieve('dataset_id') # Change the storage class to Essential dataset.storage_class = "Essential" dataset.save() |
1 2 3 4 5 6 7 | require(solvebio) # Set storage class to archive Object.update("DATASET ID", storage_class="Archive") # Set the storage class to essential Object.update("DATASET ID", storage_class="Essential") |
Supporting Archived Datasets¶
After the introduction of dataset archiving & restoring and of dataset storage classes (December 2020), a dataset may now be in an unavailable state. Scripts and apps must now check for this state before querying, or explicitly handle query failures. Both the Dataset and the Object resources now contain the "availability" parameter which returns "available", "unavailable", "restoring" or "archived" for a dataset.
See examples below:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | # Explicitly check availability datasets = vault.datasets() for dataset in datasets: if dataset.availability != 'available': print("Dataset {} availability is {}. Not querying.".format(dataset.id, dataset.availability)) continue print(dataset.query()) # Catch errors datasets = vault.datasets() for dataset in datasets: try: print(dataset.query()) except errors.SolveError: print("Dataset can not be queried: {}".format(e)) |
1 2 3 4 5 6 7 8 9 10 11 12 13 | # Explicitly check availability dataset <- Dataset.get_by_full_path("solvebio:public:/ClinVar/3.7.4-2017-01-30/Variants-GRCh37") if(dataset$availability != 'available') { print(paste("Not querying dataset", dataset$id, " with availability:", dataset$availability)) } # Catch errors tryCatch( print(Dataset.query(id = dataset$id, limit = 10, paginate = TRUE)), error=function() { print(paste("Unable to query: Dataset", dataset$id, "availability is", dataset$availability)) } ) |